Genes
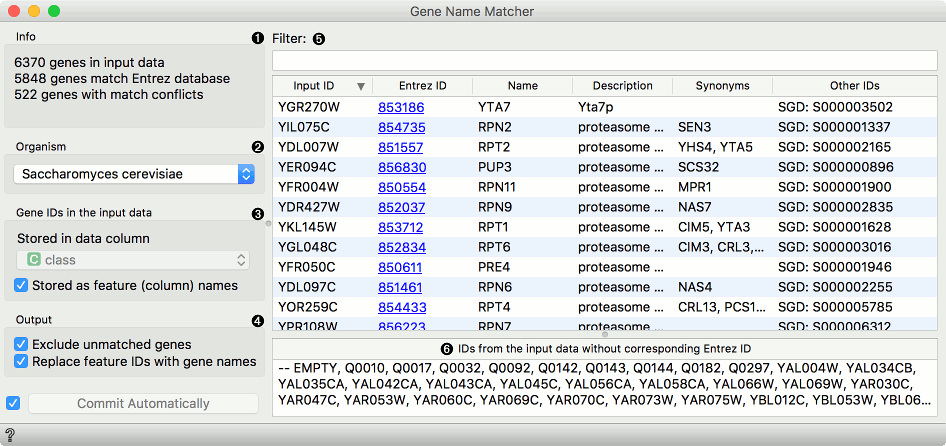
Match input gene ID's with corresponding Entrez ID's.
Inputs
- Data: Data set.
Outputs
- Data: Instances with meta data that the user has manually selected in the widget.
- Genes: All genes from the input with included gene info summary and matcher result.
To work with widgets in the bioinformatics add-on data sets must be properly annotated. We need to specify:
- Location of genes in a table (rows, columns)
- ID from the NCBI Gene database (Entrez ID)
- Organism (Taxonomy ID)
Genes is a useful widget that presents information on the genes from the NCBI Gene database and outputs annotated data table. You can also select a subset and feed it to other widgets. By clicking on the gene Entrez ID in the list, you will be taken to the NCBI site with the information on the gene.
Example
First we load brown-selected.tab (from Browse documentation data sets) with the File widget and feed our data to the Genes widget. Orange recognized the organism correctly, but we have to tell it where our gene labels are. To do this, we tick off Stored as feature (column) name and select gene attribute from the list. Then we can observe gene info provided from the NCBI Gene database. In the Data Table we can see the Entrez ID column included as a meta attribute. The data is also properly annotated (see Data Attributes section in Data Info widget).
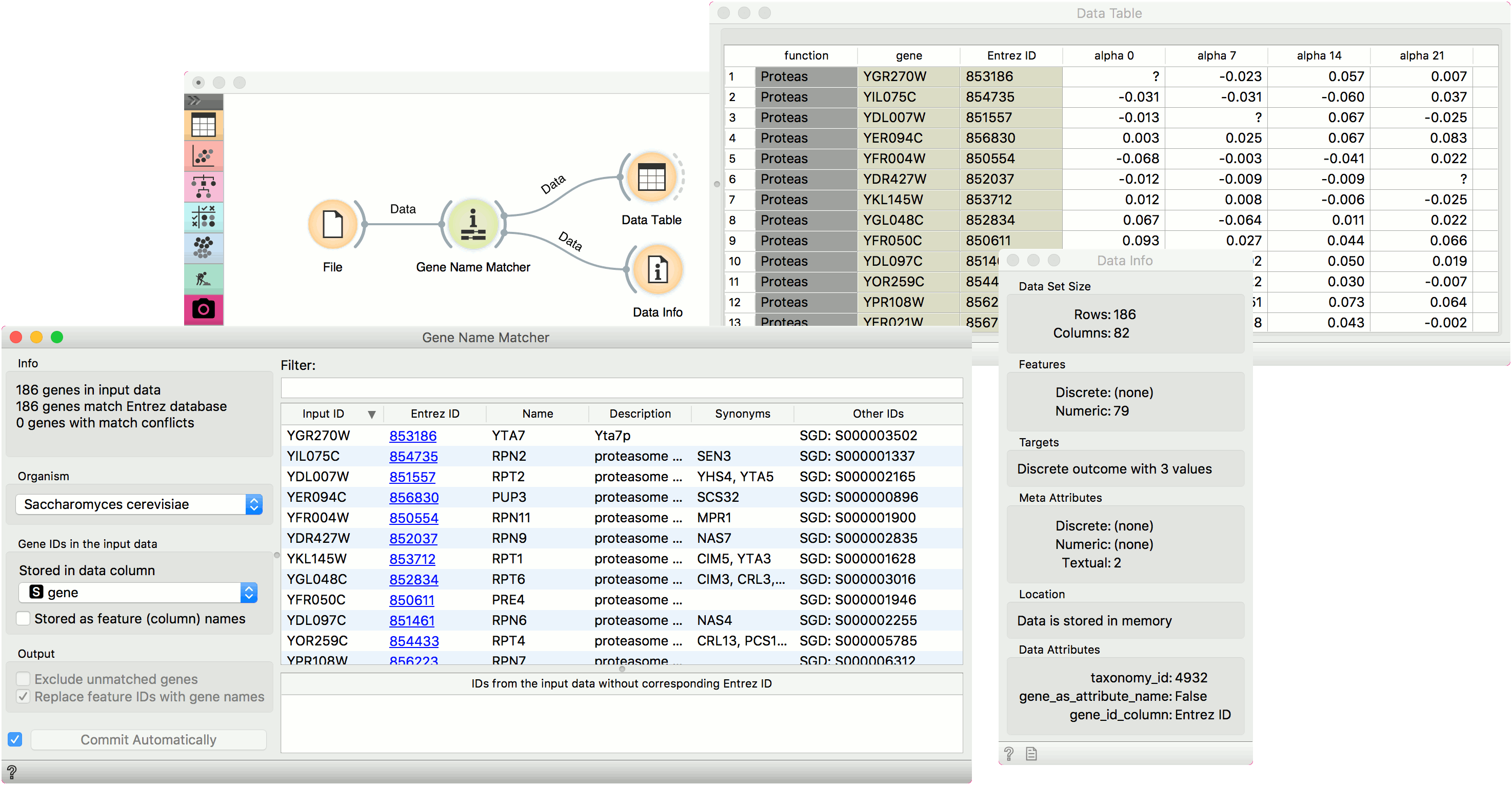
A workflow that implements this widget can be accessed here.